Why did Azelaprag fail?
A case study on using AI systems to produce holistic pharmaceutical insights
Over the past few weeks, I’ve been working on an “o1 for biotech” copilot to help understand and reason about what drugs actually do in the body. For some context, much of my background is in trying to predict how different molecules interact with the full human proteome and using this to find/design better drugs. The vastness of this data is often tedious to work with and difficult to interpret, but can be very insightful. After working with a number of labs and consulting for/cofounding multiple biotechs, it’s become clear to me that much of the space is flying in the dark when it comes to understanding how and why drugs work, which I feel someone should help with.
The problem as far as I can tell stems from how drugs are discovered: you pick a mechanism/endpoint that has evident implications in an indication, screen for the best molecules to intervene with this, and prune out/retroactively debug the molecules that don’t seem worthy of becoming drugs. Everyone in biotech knows this is grossly inefficient: thousands of molecules are pruned due to off-target effects and insufficient accession to narrowly defined mechanisms before something gets to market. Discovery is therefore decoupled from a complete understanding of how molecules are likely to behave in biological systems.
To sidestep this issue, assay developers and AI companies have been building cheaper models to predict earlier and earlier if a molecule will fail (or propose better-than-random molecules to start with). The problem with this is we are still dispensing with the holistic behavior of drugs—all models either oversimplify complex biology in vitro or rely heavily on animal/ML models that are effectively black boxes. In each of these cases, the promiscuity(/pleiotropy) intrinsic to molecules, and how this interacts with complex disease biology, is being mostly ignored or at least not explicitly interpreted, which misses critical insights that cost billions. I’m not saying do away with models; just that their inefficiency begs the question of what level is appropriate to do your modeling at so as not to sacrifice completeness and interpretability.
The second AI wave has reinvigorated visions of a future where superintelligent AI systems solve health care and cure all diseases. But are the AI drug discovery companies really building towards this, with their marginally better predictive black boxes year after year?
For the sake of this essay, I’ll focus on small molecules and protein targets as the primary currency of biotech decisions; but the thinking here is arguably useful for other modalities as well.
What if AI could run a biotech program?
To clarify, I don’t mean: “Can we create algorithms to help us find molecules/targets that perform well in some narrow predictive task?” We all know this is possible (see the last 10 years of AI drug discovery summarized in this nice post)—it’s exactly what Bioage already tried and failed to do for Azelaprag. I’m also not talking about training new foundation models on unstructured therapeutics data, since I and others have already been doing this too (if anything these are useful agents for what I’m about to describe). Neither of these really tell you dynamically why/how something works—just that an ML model said it would or wouldn’t.
Doing the thoughtful science required to run a biotech actually seems to be more of a language reasoning task than one for generic ML prediction—so long as the data to make confident decisions in biotech is available. Interpretability is key here which is why I think we need some separation between predicted/ground truth measurements and the actual reasoning core that makes and communicates recommendations. Notice how I still include predicted measurements—we can use the black boxes and other imperfect computational tools so long as we’re careful at which layer in the stack they sit. This is also why I don’t see bio foundation models as sufficient alone since they are just providing a new set of black boxes.
So the question I’m really interested in is: How can we provide the necessary tooling for an LLM to reason holistically in the same way the biotech industry does? I.e about the complete biology and mechanisms contributing to disease, what molecules might block this from multiple angles (virtually all approved drugs appear to have pleiotropy to multiple useful targets, enabling repurposing), their potential off-targets, and how to strategically build a development pipeline and IP around this? In other words, can we enable interpretability in AI drug discovery to automate the job of a full biotech company/lab?
In my own tinkering with LLMs to get them to act scientific, it seems important to prioritize chain-of-thought (CoT) and multi-agent reasoning with access to disease biology, patent literature, and chemical databases. What I think has been missing (which seems evident when I help biotechs understand their own molecules) is an adequate bridge between the relevant biology and chemistry. That is the keystone between drug structure ⇄ function interpretability is the complete biochemical interaction data. While lots of biochemical assays have been done over the past few decades, protein interaction data available to current models is fairly sparse considering there are 20,000+ proteins in the human body and at least 37 billion commercially available small molecules (not counting other modalities). If we could provide this rich interaction data on the fly, even if approximated, LLMs might be able to bridge the gap and holistically intuit how molecules affect entire biological and clinical systems.
I’ll comment on how I was able to gather a useful approximation for this data at the end of this post. Just know for now, I essentially use a combination of molecular docking and ligand similarity analyses that score compound-protein interactions from 0-1. I then built a copilot during my finals last semester to help manage and parse this context for a set of 13k compounds ⨯ 23k proteins. Here’s a breakdown of a conversation I had with it where I tried to understand Bioage’s recent bust, Azelaprag…
Holistic insights on Azelaprag interactions in the body
Azelaprag (BGE-105) was Bioage’s flagship program to prevent muscle atrophy when taken with GLP-1s. Some helpful context to know:
Bioage reported APJ agonism as Azelaprag’s mechanism of action.
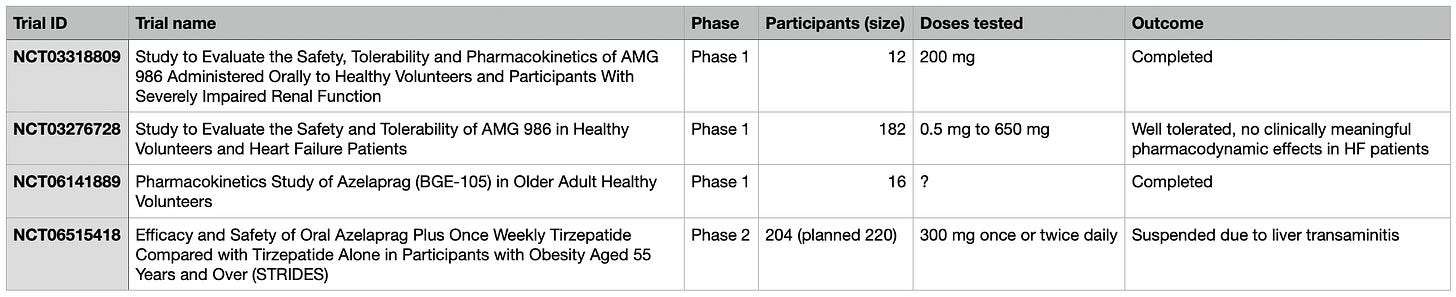
Azelaprag showed apparent human tolerability in prior Phase 1 trials for doses up to 650mg (200mg in the only completed trial)1.
Bioage dosed Azelaprag at 300mg in Phase 2.
$500M raised for a sexy proteomics platform and an IPO later, Bioage’s flagship flops in Phase 2 due to signs of liver toxicity (2024 Q4). I was bullish at first. So I tasked my copilot to help understand what happened:
APJ binding
First, I wondered if Azelaprag was even a potent APJ binder. Why? I was curious about the dose selection and the best information I could find about the potency of the APJ interaction was from this paper, where only functional measurements suggested APJ was being agonized by Azelaprag. The patent disclosure didn’t offer much on direct binding affinity either, only adding experiments looking at expression. I tend to think in terms of free energies so this wasn’t helping my intuition. Nonetheless, Bioage says in its Phase 1b press release that Azelaprag binds APJ, so we’ll run with this for now. Given that agonists known to directly bind (e.g. apelin itself) can result in increased downstream APJ expression, this could explain how Azalaprag induces APJ expression too.

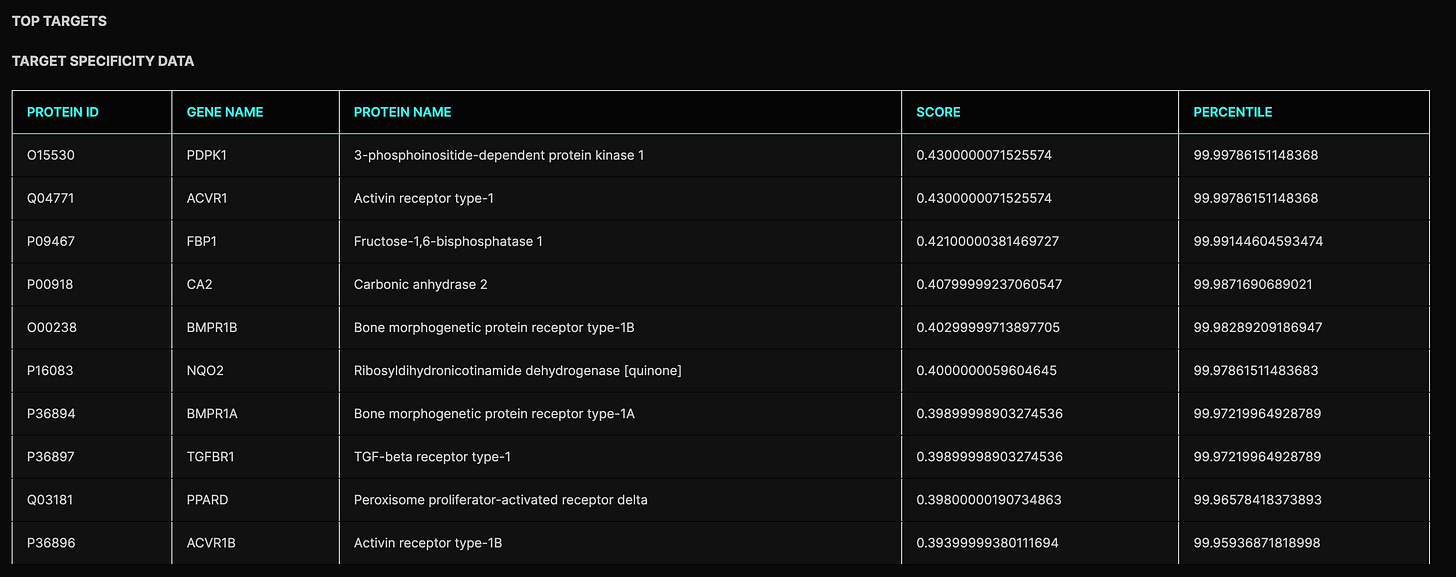
A few interesting insights from the copilot right off the bat. First, the docking here doesn’t seem to suggest anything definitive on whether or not APJ is a potent binding partner of the drug. The APJ interaction happens in the ~90th percentile relative to the rest of the proteome, so fairly high at first glance. On the flip side, the 90th percentile corresponds to about 2000 stronger predicted interactions—not exactly what we’d expect given this is the reported mechanism. Since the scoring relies heavily on comparisons to previously associated ligands, this could just be a reflection of the relative novelty of Azelaprag as an APJ agonist. This certainly seems true when looking at APJ ligands in the PDBe-KB. In that case, the interaction may be potent in reality with this data merely suggesting other strong, unaccounted interactions. More interestingly though, Azelaprag doesn’t even seem remotely competitive for APJ relative to other approved/experimental drugs. Assuming that the ligands associated with APJ in the scoring system do carry some generalizable information about the chemical space inhabited by true binders, which seems narrowly true from the 90th percentile stat, it’s not so clear what’s going on here. All we can say is this prior doesn’t seem to conclusively give us a model of competitive APJ-Azelaprag binding.
What if direct agonism wasn’t as reliable a conclusion as the functional assays led us to believe? I wouldn’t be so quick to throw away the positive assay data in favor of an in silico non-result. Personally, I trust the functional assays (especially given competitiveness with apelin-13) and Bioage may even have corroborative binding data. If anything, the modeling above might suggest other kinetic factors, i.e. other interactions (possibly even playing a role in bioavailability), detracted from highly specific APJ binding in vivo. If we take the not-so-definitive docking at face value, this could explain the choice to increase dosing from 200mg and subsequent toxicity with tirzepatide. That is naively, indefinite docking results could be reflective of low or context-dependent target selectivity, which may have led to off-target effects and poor tolerability. This does align with what we saw in STRIDES.
Other predicted interactions
A very pressing issue for me at this point was: if the mechanism wasn’t selective by binding, how could Bioage see 1) elevated APJ levels and 2) muscle preservation in the first place? For the record, I don’t think Bioage was intentionally falsifying results. The simplest explanation was that they found the best clinically safe + in-licensable APJ agonist they could, and got lucky with other more potent interactions to show functional efficacy in muscle preservation models. I think getting lucky with helpful pleiotropy is more common than we realize in drug development but I’ll save this for another essay (hint: if all small molecules are promiscuous then the ones that become drugs may just be promiscuous to non-toxic things).
To stay focused though, I then prompted my copilot to unpack off-target predictions:


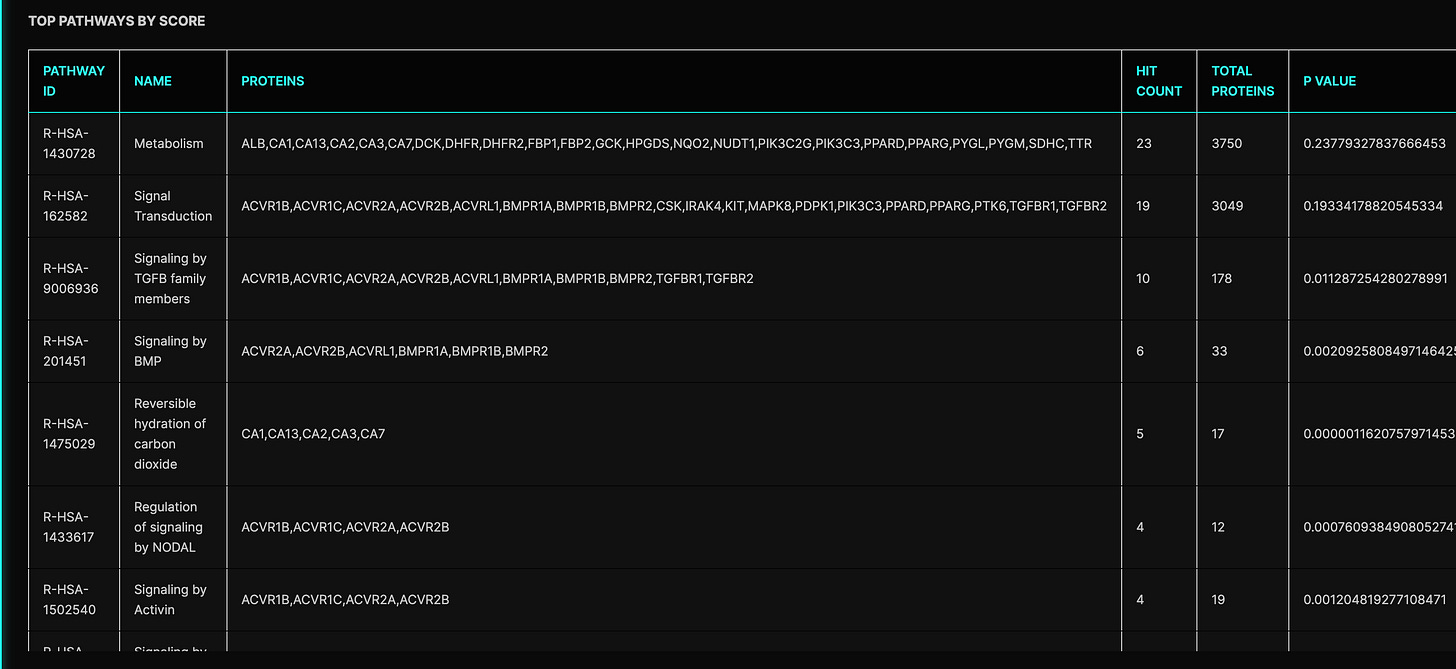
Some of these top interactions actually looked pretty interesting. For example, TGF-beta, PDK1, and Activin receptors are all very well-implicated in muscle growth and maintenance. Moreover, Figure 4 in the patent disclosure shows that Azelaprag increased Akt phosphorylation which is known to occur via PDK1, so we have a possible mechanistic corroboration.
Given these predicted interactions and the preclinical models used by Bioage, it’s not inconceivable that without potent APJ binding, Azelaprag may have shown protection against muscle wasting via interactions with TGF-beta, Activin, etc. To make this even more interesting, TGF-beta has been shown to inhibit apelin production which we said earlier can also contribute to APJ expression. So there could have been a multifaceted effect here if Azelaprag was disinhibiting APJ expression via TGF-beta.
So then what caused the failure? If anything it seems like these interactions might offer some serendipitous pleiotropy, or at least explain the increased APJ expression seen without strong agonist binding…
The chat logs indicate wariness by the copilot for these interactions too. For example, diverse TGF-beta pathway members were highly enriched in the top targets analysis conducted by the copilot. Several times the bot noted this as a cause for concern. Given that the STRIDES trial was shut down due to signs of liver toxicity at the 300mg dose, one plausible story emerges: APJ was a safe target but Azelaprag had insufficient direct agonism, with observed efficacy explained by interactions with dodgy upstream regulators. This could directly suggest possible off-targets to test. For example, TGF-beta signaling is highly context-sensitive and pleiotropic in the liver with known implications in hepatotoxicity. Context sensitivity may explain why signs of liver injury were present but not so widespread in the trial (and went undetected in Phase 1—although how hard were clinicians looking for transaminases in Phase 1?). That is, if TGF-beta interactions are only toxic in the liver under certain, non-homogenous conditions, it’s not impossible that we were simply unlucky in not picking up on this earlier. An important caveat here is this explanation relies on elevated transaminases really not being seen prior in Phase 1, i.e. Amgen didn’t just have this data and chose not to report it.
To complicate this further, tirzepatide itself could have synergized poorly with Azelaprag to cause toxicity. It’s known that the tirzepatide-only arm of STRIDES saw no warning signs but we know drug-drug interactions can be dodgy. It was difficult to find compelling explicit connections between GLP-1 biology and the major Azelaprag interactions I predicted, but this very well could have been problematic. Further investigations seem warranted in checking 1) where metabolic Azelaprag interactions may have blocked or synergized poorly with GLP-1 biology and 2) where downstream GLP-1 effects may have synergized specifically with notable liver-implicated Azelaprag interactions. Hopefully, the predictions here can clue us into experiments we can run to verify where toxic synergies may have occurred.
The model crawled through a lot of literature that I haven’t properly cited so I’m including a file link to the copilot scholarship here.


Alternative MoAs or off-targets?
So was this the off-target toxicity that caused the trial to fail? There is still something unsatisfying here. I’m skeptical that the failure is as simple as this considering a lot of the predicted “off-targets” also happen to be implicated in the primary endpoints. The copilot also notes that even though some of these pathways may cause liver injury, there isn’t much direct causal evidence for roles in transaminitis, which is the more specific reason STRIDES was suspended.
So maybe these interactions were okay and the reported mechanism was just more downstream of true Azelaprag binding partners than previously thought. Then shouldn’t there still be some therapeutic window, even if some of these predicted targets are scary at high doses? After all, basically every drug target becomes toxic at high enough concentrations. I assume Bioage may have based their decision to go with 300mg/day (instead of a proven 200mg which was safe and seemingly effective for another program) not only on insufficient APJ agonism but also that Azelaprag had lower-than-desired potency in functional models. After all, even if 650mg was reported safe in Phase 1, why would you gamble with increasing the dose from a minimal yet effective 200mg unless you needed to for the new use case? I.e. wouldn’t you generally want to play it safe in the lower end of your therapeutic window? Since biotechs tend to be more conservative with increasing doses than decreasing, I’m not ruling potency issues out as a culprit.
Could we have then known that low potency and a subsequent need to increase dose would be an issue? Typically in drug development, low potency to a mechanism is only a real problem if the drug isn’t selective. So then the question becomes: Is Azelaprag selective to any of these other possible mechanisms? There was a key insight above that I (and my copilot) glossed over…
While many muscle preservation targets were competitive to Azelaprag relative to other proteins, Azelaprag wasn’t competitive to any protein relative to the rest of my approved + experimental drug library (competitive defined as top 100 binders out of 13k molecules). Typically to be certain of strong binding, I’d like to see drug mechanisms show up both as top predicted targets of the compound as well as in protein lists for which the drug outcompetes other compounds. The latter did not happen here. I assume this means Azelaprag mechanisms were not potent enough to result in a clean drug with well-controlled off-target interactions (assuming there are off-targets here other than TGF-beta, Activin, etc), despite its broad interaction profile being biased towards potentially useful pathways.
Not included in the screenshots above were summary statistics on broader Azelaprag interactions that the copilot retrieved: thousands of proteins had non-zero interaction scores. Does this automatically imply a dangerously messy compound? Probably not. When scoring across the proteome, this really isn’t surprising given the number of models we’re running. Pretty much all drugs seemingly have a similar level of messiness built in. Small molecules are nimble and kinetically attempt interactions with nearly everything they bump into; the scoring models seem to simply be recovering this. What mitigates the predicted messiness in successful drugs seems to be extremely competitive interactions happening bidirectionally from [compound → protein] and [protein → compound]. I intuit this as the drug not only outcompeting other drugs/natural ligands for the target; but also the target outcompeting other proteins for the drug. If this doesn’t happen for a drug and some set of disease-implicated targets, it seems 1) the drug tends to not be efficacious, and 2) those other non-zero interactions have real consequences at high concentrations.
So why do I think the trial failed? The tl;dr is: Azelaprag actually does seem messy. Did you need to read this essay to know that? No. Knowing it failed due to signs of toxicity would have sufficed to reach that conclusion. Whether the messiness manifested itself simply as hard-to-detect off-targets, or whether these interactions were somehow uniquely problematic in combination with tirzepatide, is still an open question. But how did Azelaprag manage to convince Bioage and us it would work despite its messiness? To summarize the complete thinking my copilot helped flesh out about why Azelaprag may have been tricky:
Despite Azelaprag being the best APJ agonist Bioage found, the interaction may have not been highly competitive.
Other strong protein interactions of Azelaprag could have helped achieve preclinical efficacy even in the absence of competitive APJ binding.
Azelaprag simply could have been toxic from these off-targets in combination with tirzepatide at 300mg, but maybe they were actually useful.
While seemingly useful targets were the strongest predicted binding partners of Azelaprag, none were particularly potent relative to what we’d expect of a typical drug, closing the therapeutic window in combination and opening the door to other off-target toxicity.
Was this avoidable? That’s for you to decide. To Bioage’s credit, knowing where to speculate failure early on in diligence is a hard unsolved problem and a big motivator for building this copilot. But to harp on a point I made at the beginning of this essay: black-boxed AI/in vivo models seldom on their own offer the biochemical interpretability to say why/how a drug might work. More care can always be taken before placing big bets.
Caveats
In the absence of assayed biochemical data for Azelaprag, I (and the LLM that helped me) had to look for clues in extensive simulated data. As someone who spends lots of time with molecular docking, I’d be the first to say we should be careful with these results. Docking can be incredibly insightful but can also easily mislead us.
To provide some intuition on the docking method, so you can decide how trustworthy these results are, the copilot (via API) scores unmeasured interactions between protein and new query ligands by comparing said ligands to known binders of the target. While fairly simple, this actually feels close to what nature does when docking molecules to proteins. Most druggable binding pockets evolved for some set of natural ligands, so the problem of docking new drugs may be thought of as querying structural similarity to these known ligands. Now, this isn’t preciseley true (e.g. allosteric binding pockets and structurally novel inhibitors do exist) and in fact the protocol here doesn’t confine itself to natural ligands/known binding pockets, but I’ve found the intuition is helpful to understand what the interaction scores actually mean. That is, they reflect compound similarity to the best information we have available on each target’s natural, synthetic, and high-confidence predicted ligands.
For those less interested in intuitions, the scoring protocol leveraged here has been used extensively before to make novel therapeutic predictions that empirically work, and somewhat uniquely gets more reliable when you string together the results of many calculations across the proteome (e.g. above: maybe a high interaction score to ALK5 alone isn’t convincing, but high scores to multiple homologous receptors in the TGF-beta pathway warrants some further investigation). We also know molecular docking more generally (beyond methods used here) works best—even for finding new binders—when known ligands are factored into the prior of the system. This is maybe part of what drives performance in AF3/Diffdock.
As I’ve been building the copilot, I’ve assumed the important thing at the earliest stage in biotech decision-making is not so much “What narrow evidence adds to my conviction in working on this asset?” (which classical ML/bio FMs help with) so much as it is “How do I interpret the market/scientific landscape to know early on where an asset might fail? (And can I flip this to find real alpha?)” While I’d argue autonomous biotech copilots could systematize this by digesting and reasoning with all available data and solve the “cold-start” problem of launching a new venture/program—which is really a function of who has the most comprehensive intuition about what a molecule or target might be useful for—the current reasoning engine is far from complete. I’m optimistic that I’ll be able to continue improving this to be more autonomously thoughtful; but the fact that it isn’t writing this post for me yet is disappointing. Even in its current form though, I see myself using it to speed up ongoing projects/collaborations. I think of its current offerings as a high-throughput way to screen full biotech plays instead of just compounds.
This holistic reasoning endpoint feels like a much more tangible step towards the AGI healthcare utopia that AI proponents have been imagining. But how far off are we really from this? Again, I don’t think bio foundation models like PreciousGPT/etc are offering much on interpretability given that so much still happens under the ML hood. A few players have started to emerge that seem more aligned toward truly automating science (Future House, Mithrl), but as far as I can tell, they don’t yet offer anything much different than what’s afforded by Cursor and Perplexity. That said, they are evidence that the tech stack to automate biotech is growing.
Final thoughts and things I’d like to see done
Even if you choose to take this data seriously, I’d like to emphasize that for me, this essay is more of a proof-of-concept that you can get LLMs to 1) manage context and help reason effectively about omics-scale information without new foundation models, and 2) provide intuitions (not concrete results) for where to look/what assays to run to better understand poorly characterized compounds, like Azelaprag.
So as a first call to action, someone please go check on Azelaprag interactions with TGF-beta family members/etc. If anyone has the APJ binding data, this would also be great to see—I couldn’t find this on my own. I’m sure there is plenty to learn so hopefully we can prevent failures like this from happening again.
Second, there are way more insights baked into this model than I can ever hope to take advantage of on my own. I’m personally interested in aging and addiction and will be seeing if I can tease out any new leads for assets to work on there, but please feel free to reach out if you’re trying to do diligence on a biotech play and want help—average runtime for the copilot is ~5 mins and these conversations are a lot of fun for me.
Third, I’m looking for ways to effectively recreate this compound-protein interaction database in the lab. This seems important for 1) confirming the insights this setup already produces and 2) offering more ground truth data for my system to parse (+ fine-tune drug design models with). So if you are or know of someone thinking about large-scale or multiplex binding arrays/high-throughput protein purification, please reach out. I’m amassing a network of people in this headspace and would love to add more people to it.
Finally, assuming this simulated data on Azelaprag holds up, I don’t think the takeaway from this exercise is: “If Azelaprag was more selective to its target, none of this would have happened—another win for lock-and-key drug discovery!” If Azelaprag’s “off-targets” weren’t highly enriched in muscle preservation pathways, I’d be on board with this conclusion. However, Azelaprag showed efficacy for a functional endpoint that was actually implicated with its predicted off-targets, which seems like a near-miracle and worth investigating more. If anything, we should actually use this as an indicator that looking for useful pleiotropy has a ton of potential if we can ensure better command over true off-targets and therapeutic windows. It also might be the key to an aging trial.
If you have thoughts on any of this or just want to chat, you can reach me at my personal or school emails. I look forward to it and keep an eye out for future writings.
Acknowledgments
The data generated here would not have been possible without Ram and the rest of the CANDO team. I’d also like to credit some friends in Norn: Satvik for pushing me to write this; and Marton for good conversations about Bioage+biobanks+in-licensing which helped refine my thinking here. Lastly, I was partly inspired to build this biotech copilot after seeing a friend of mine, Ron, do something similar for financial data, so definitely check him out too if interested in these kinds of AI systems.